Computational and Experimental Discovery of a Conformational Switch Controlling Replication and Translation in a Plus-Strand RNA of the Turnip Crinkle Virus

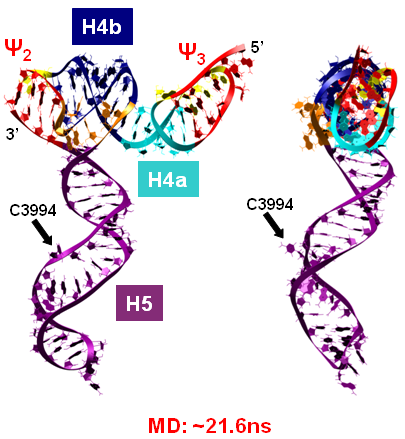
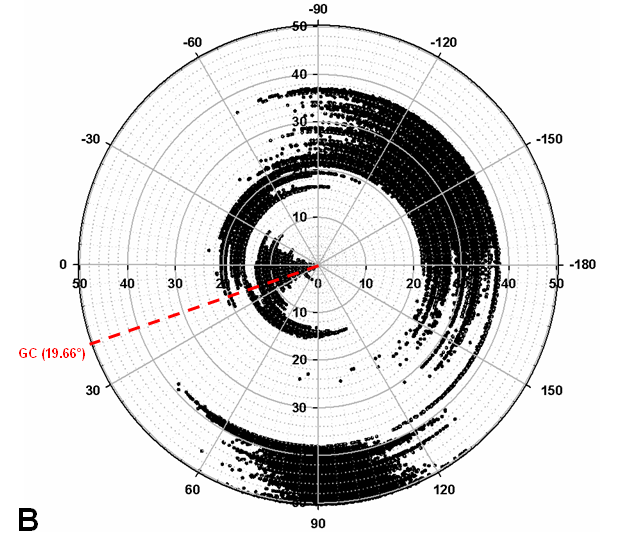
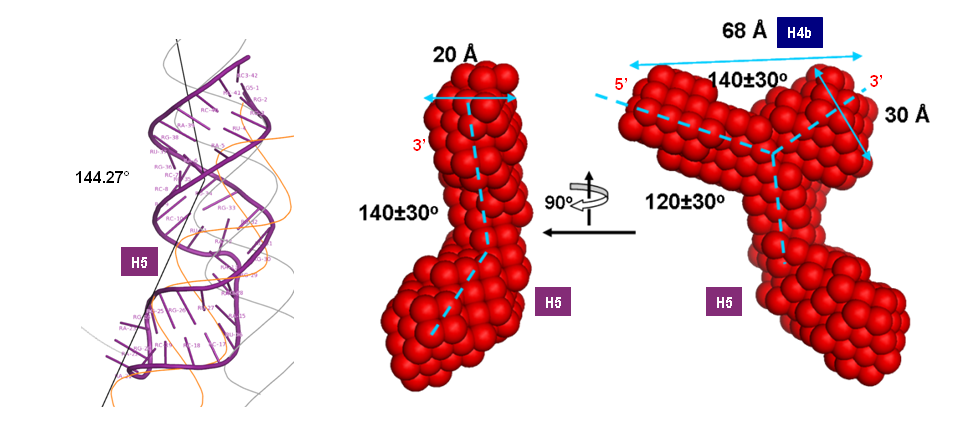
Turnip Crinkle Virus (TCV) is one of the smallest known RNA plant viruses and therefore represents an excellent candidate for study to understand the structural and functional basis for its replication and translation. TCV genomic template (see Fig 1) is neither 5'-capped nor polyadenalated. It was known that TCV contained a translational enhancer within its 3’ UTR, but how this enhancer worked was unknown. By combining computational and experimental methods, we elucidated the secondary and 3D structure of the translational enhancer (see Fig. 2). Our massively parallel genetic algorithm, MPGAfold, was used to predict the secondary structure of the 3' terminal 195 nt region (Zhang et al. 2006). Compensatory mutagenesis analyses in vivo and in vitro in-line structure probing confirmed the existence of the key predicted features (stem-loop motifs an one H-type pseudoknot - Ψ3) and added another pseudoknot to the model – Ψ2 (see Fig 2). 3D modeling utilizing our own package RNA2D3D (Martinez et al. 2006) produced a structure resembling a tRNA (Fig 2) (McCormack et al. 2008). Thus the existence of the first internal tRNA-shaped structure (TSS) was established. Molecular dynamics simulations (MD) in explicit solvent showed the stability of the entire modeled structure (100nt) and indicated high flexibility of some of its elements, exemplified by an outstanding rotational mobility of C3994 (see Fig. 3), in agreement with the in-line structure probing data (McCormack et al. 2008). In addition, MD simulation of just the H5 stem-loop (42 nt) extracted from the model of the full TSS was performed. Using the data from the entire trajectory and the most stable interval of the MD simulation (the last 18 ns), we measured the angle between the proximal and distal helices withing the H5 stem-loop. These match the recently solved TSS structure in solvent. A novel protocol combining Small Angle X-ray Scattering and Residual Dipolar Coupling (SAXS/RDC) data (Wang et al. 2009; Xiaobing et al. 2010) was used in this study. The overall shape of the TCV translational enhancer was confirmed (see Fig. 4), and the details of the relative orientations between the TSS elements were elucidated (Xiaobing et al. 2010).
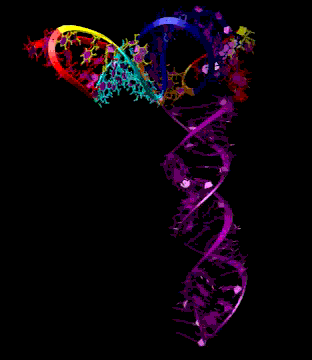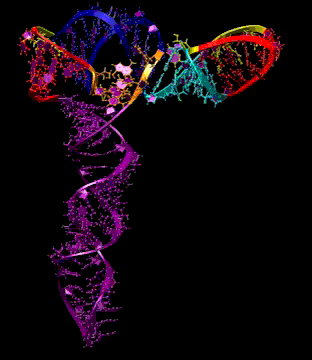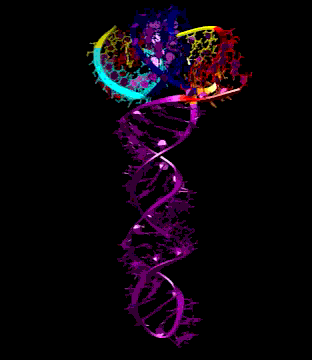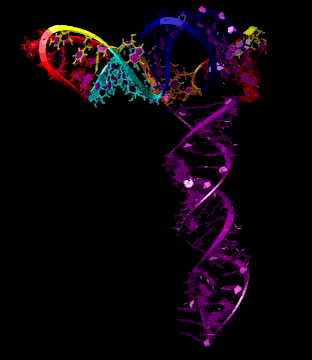
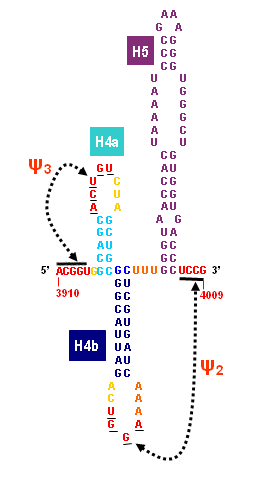
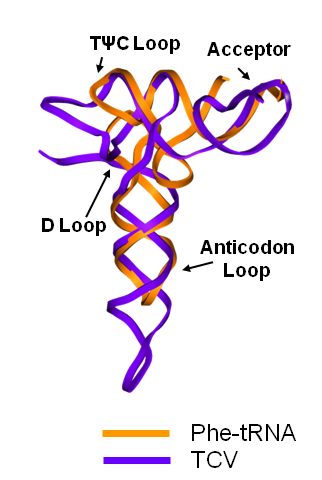
The structure-implied functionality of the TSS has led to further experiments aiming to determine how the interconnected 3' UTR sequence and structure elements participate in the processes of translation and replication. The shape of the modeled structure suggested that the translational enhancer may potentially act by binding ribosomes. Indeed, the TSS was experimentally found to bind the 80S ribosome the 60S large ribosomal subunit (Stupina et al. 2008). Further experiments showed that the RNA dependent RNA polymerase (RdRp) also binds in the vicinity of the enhancer element. This interaction appears to induce a conformational change in the element and some surrounding context, which inhibits further ribosome binding and leads to initiation of the transcription of the negative strand. RdRp binding appears to be concentration dependent, and the conformational change it induces is reversible (Yuan et al. 2009).
Thus the series of computational and experimental discoveries has led to the finding of a new type of translational enhancer element. It is new from a structural point of view, since the tRNA-shaped structure (TSS) is not a tRNA on a secondary structural level. It is the first such element found within a 3' UTR, and it is part of a newly found conformational switch. The mechanism described in (Yuan et al. 2009) indicates how translation and transcription are controlled in this virus. One could argue that similar mechanisms may exist in other eukaryotic genomes.